scVDJen
scVDJ-seq User Manual
Chapter 1 Scope of Application
Single-cell immune repertoire analysis (hereinafter referred to as scVDJ-seq) is a technology used to study the gene expression and immune response of individual immune cells. The following kits can handle high-throughput sequencing data analysis:
DNBelab C Series High Throughput Single Cell 5'RNA & V(D)J library preparation kit set.
Chapter 2 Product Introduction
2.1 Analysis flow chart
scVDJ-seq analysis process is an automatic analysis process developed based on DCS cloud platform system, which includes 5-terminal RNA analysis, data quality control, VDJ library reads extraction, de novo assembly and annotation, effective cell screening, Clonotype identification and report generation:
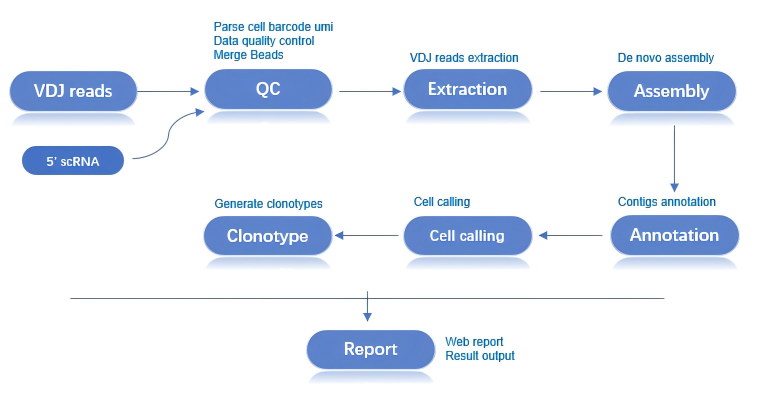
2.1.1 5-terminal RNA analysis
The content is the same as the scRNA-seq_v3 analysis, the difference is only that the scRNA-seq_v3 is analyzed from the 3 end, scVDJ-seq from the 5 end, to find the corresponding relationship between cells and magnetic beads.
2.1.2 Data Quality Control
By identifying the corresponding relationship between the cell barcode and the cell, the sequencing data is separated into single cells through data quality control and magnetic beads.
2.1.3 Get VDJ region sequence
The RNA data of each cell was aligned with the reference sequence of the VDJ gene using TRUST4, and the VDJ region sequence was extracted.
2.1.4 VDJ de novo assembly and library annotation
The VDJ region of each cell was De Novo assembled and library annotated using trust4.
2.1.5 Effective cell screening and Clonotype identification
According to the results of VDJ assembly annotation and the cell acquisition of the 5-terminal transcriptome, cell filtration is carried out, and the effective cells obtained are identified by Clonotype.
2.1.6 Report Generation
Comprehensive analysis of the results, collated into HTML report.
Chapter III Instructions for Use
The scVDJ-seq analysis process uses the DCS cloud platform system to manage the whole process of sample input and report output. The following section describes how to use the scVDJ-seq analysis process based on the DCS cloud platform system.
3.1 Summary
This chapter describes how to use the scVDJ-seq analysis process for analysis. Before using it, please read and understand the contents carefully to ensure that the scVDJ-seq analysis process can be used correctly.
3.2 Use Scenario 1: manual delivery
The operation includes four steps: uploading data, constructing a reference genome (optional), inputting sample information, and initiating analysis. After completing the sample information entry, run the task. When the customer sees that the task status is![]() When completed, it means that the task has been completed and the report section can be viewed (see3.4 Part)。
When completed, it means that the task has been completed and the report section can be viewed (see3.4 Part)。
3.2.1 Step 1: Upload Data
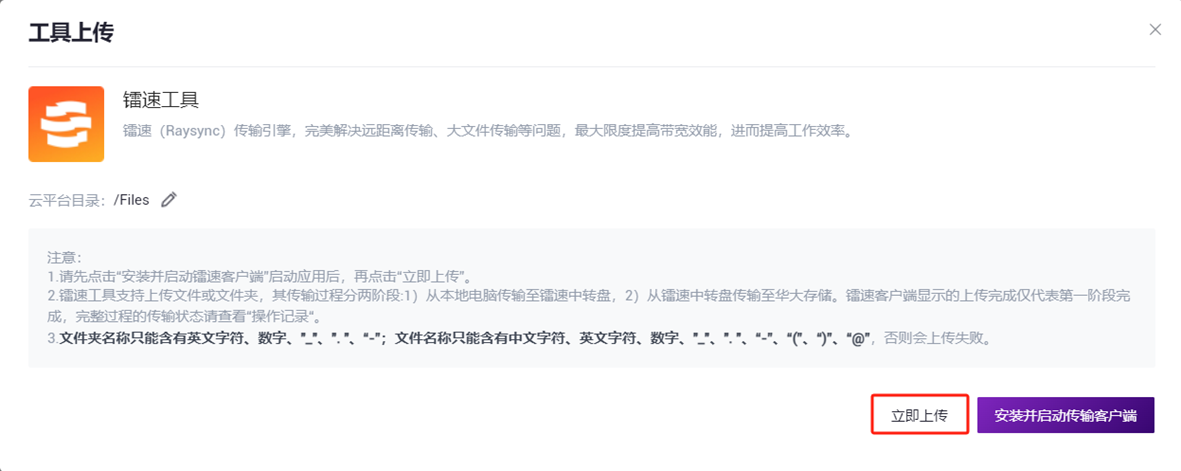
Click [Data] in the navigation bar to enter the Data management page, enter the target folder, and click [+ Add files]-[Tool upload] in the upper right corner to upload Data (Figure 3-1):
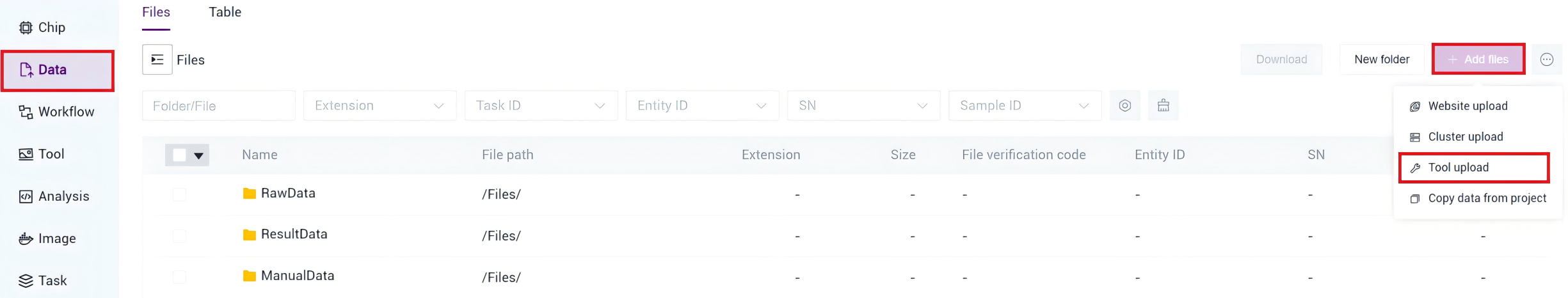
Figure 3-1 File Upload Step 1 Click [Upload] to browse and select the required files (Figure 3-2). After uploading, the files will be displayed in the target folder (if uploading for the first time, you need to click [Install and start transport client] to Install the required tools):
3.2.2 Step 2: Build a reference genome (optional)
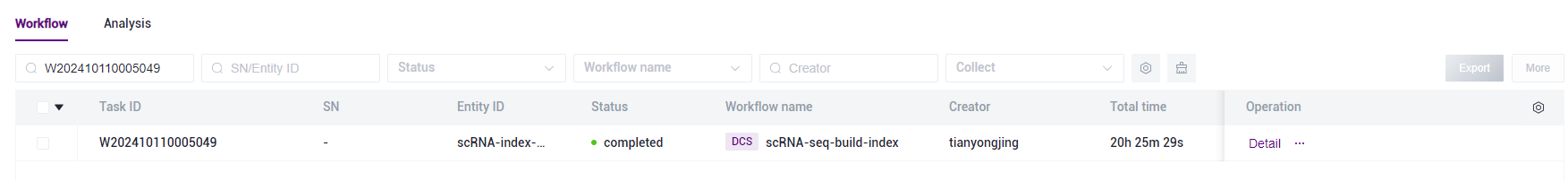
Click [Workflow] in the navigation bar to enter the process analysis page, enter scRNA-seq-build-index in the search box, and click [Run] (Figure 3-3)

Figure 3-3 Reference Genome Construction Step 1 Select Run workflow, enter Entity ID, and click [Next] (Figure 3-4):

Figure 3-4 Reference Genome Construction Step 2 Enter the reference genome information and click [Next] (Figure 3-5):

Figure 3-5 Reference Genome Construction Step 3
Reference genome information input parameter description:
refName: The species name of the reference genome, which is used for report Display;
GTF: Genome annotation GTF file, its chromosome name needs to be sorted and consistent with FASTA file, does not support compression format;
chrM: name of mitochondria;
FASTA: a genome FASTA file. The chromosome name must be the same as that of a GTF file. Compression format is not supported;
Cpu: CPU required for operation;
Mem: Memory size required for operation.
Click [Run] to start the analysis (Figure 3-6):

Figure 3-6 Reference Genome Construction Step 4 After the task is completed, the Status is displayed
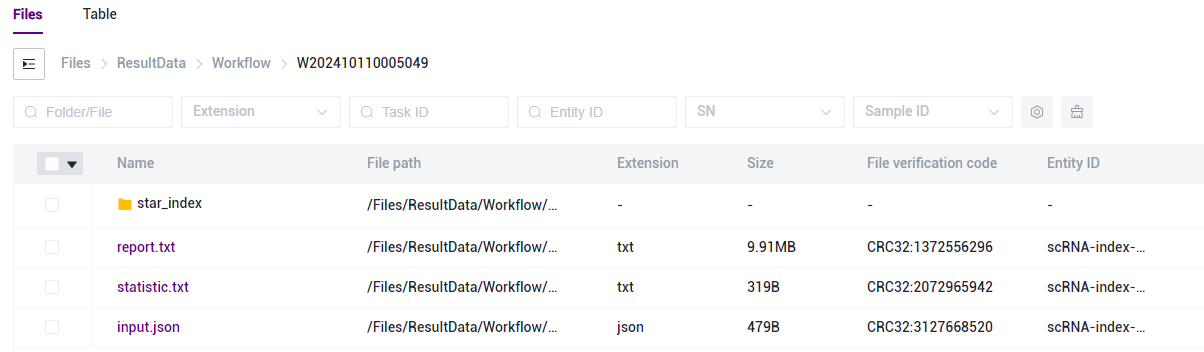
 completed, copy the Task ID (Figure 3-7), click the navigation bar [Data], and enter the Task ID to search, where the star_index file will be used as the reference genome file to participate in the 5-end RNA analysis module in the scVDJ-seq analysis (Figure 3-8).
completed, copy the Task ID (Figure 3-7), click the navigation bar [Data], and enter the Task ID to search, where the star_index file will be used as the reference genome file to participate in the 5-end RNA analysis module in the scVDJ-seq analysis (Figure 3-8).
3.2.3 Step 3: Sample Information Entry
Click [Workflow] in the navigation bar to enter the process analysis page, enter scVDJ-seq in the search box, and click [Run] (Figure 3-10):
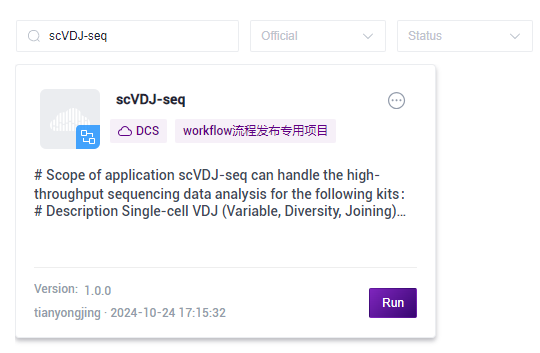
Figure 3-10 Sample Information Entry Step 1 Select Run workflow, enter Entity ID, and click [Next] (Figure 3-11):
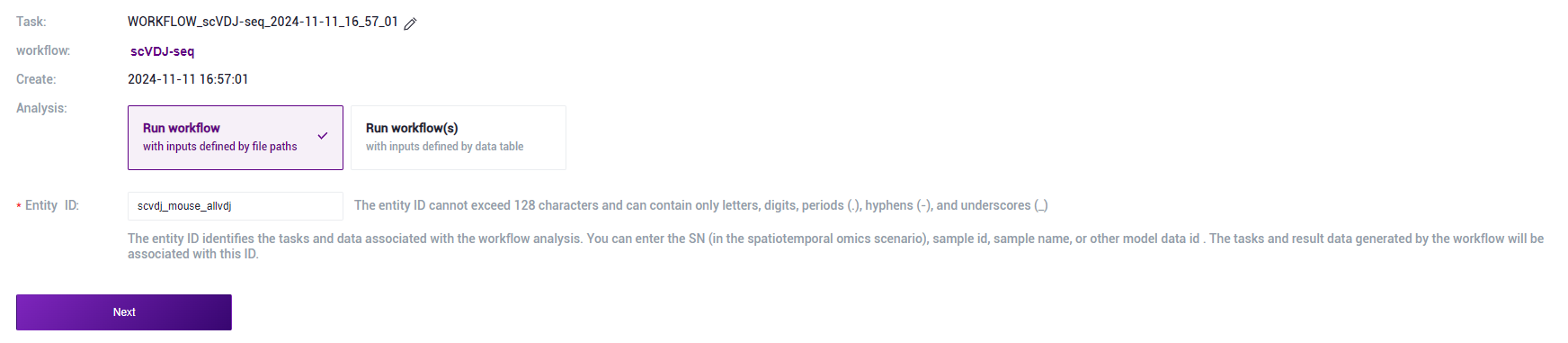
Figure 3-11 Sample Information Entry Step 2 Enter the sample information and click [Next] (Figure 3-12):
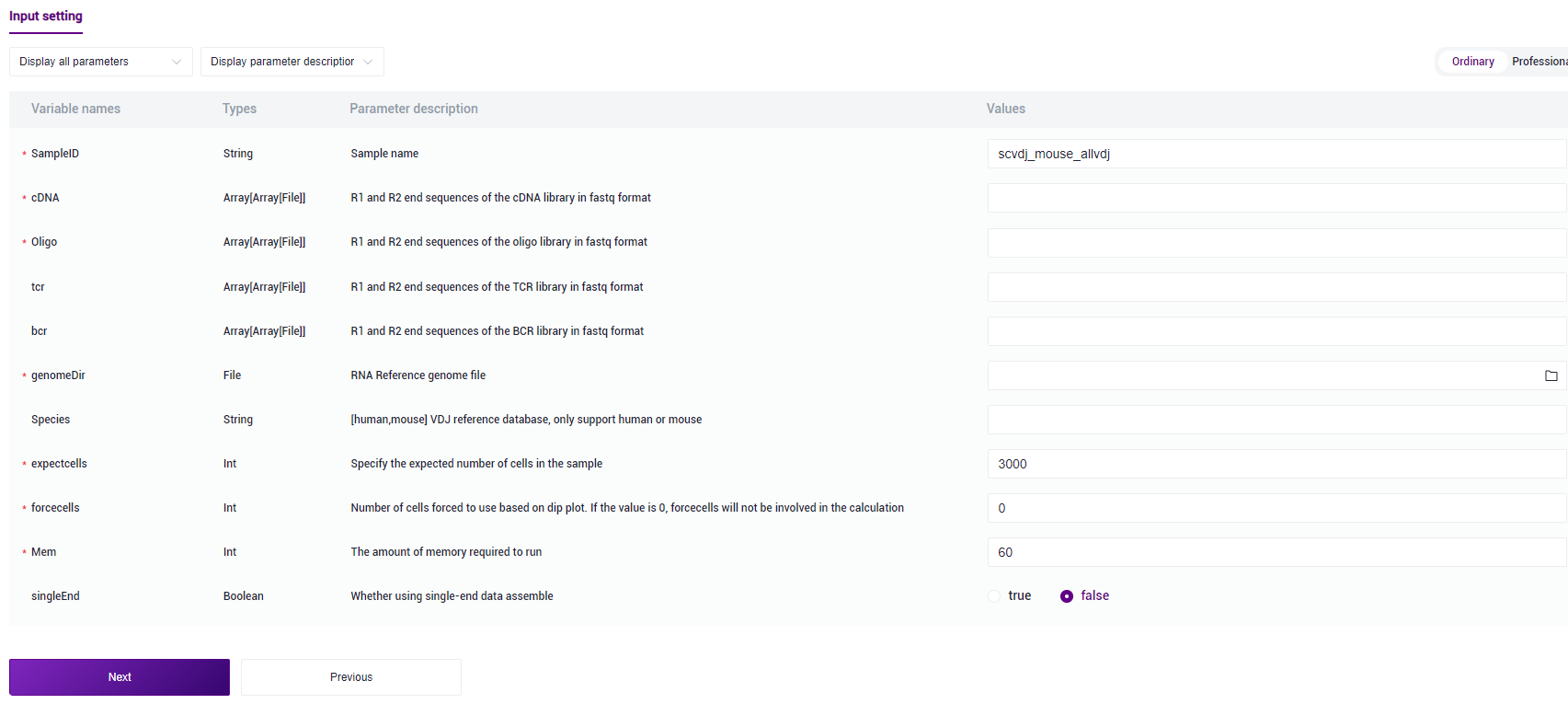
Sample information entry parameter description:
SampleID: the name of the sample, which is the same as the Entity ID by default;
cDNA: R1 and R2 end sequences in fastq format of cDNA library;
Oligo: the R1 and R2 end sequences of the oligo Library in fastq format;
tcr: Optional. R1 and R2 end sequences of tcr Library in fastq format. If not input, TCR library will not be analyzed;
bcr: Optional. bcr Library fastq format R1, R2 end sequences, if not input, do not carry out BCR Library analysis;
genomeDir: reference genome file for 5-terminal RNA analysis. For example, in step 2 (3.2.2 Construction of reference genome), the star-index file in the Task ID folder is selected;
Species: the reference database for VDJ analysis. It only supports human or mouse. Only human or mouse can be filled in. If other strings are filled in, TCR and BCR Library analysis will not be performed. If tcr or bcr files are not entered, this parameter will not take effect and may not be filled in;
expectcells: the desired number of cells, the number of cells can be selected according to experience;
forcecells: force the number of beads to be selected, and force the number of beads to be intercepted according to the sudden drop chart; If it is 0, it will not participate in the calculation and retain all cells;
mem: memory size required for running. The default value is 60g. Running VDJ analysis requires a large amount of running memory, and it is recommended to set at least 100g.
singleEnd: indicates whether to use single-ended data Assembly. The default value is not used.
3.2.4 Step 4: Start Analysis
Click [Run] to start the analysis (Figure 3-13):
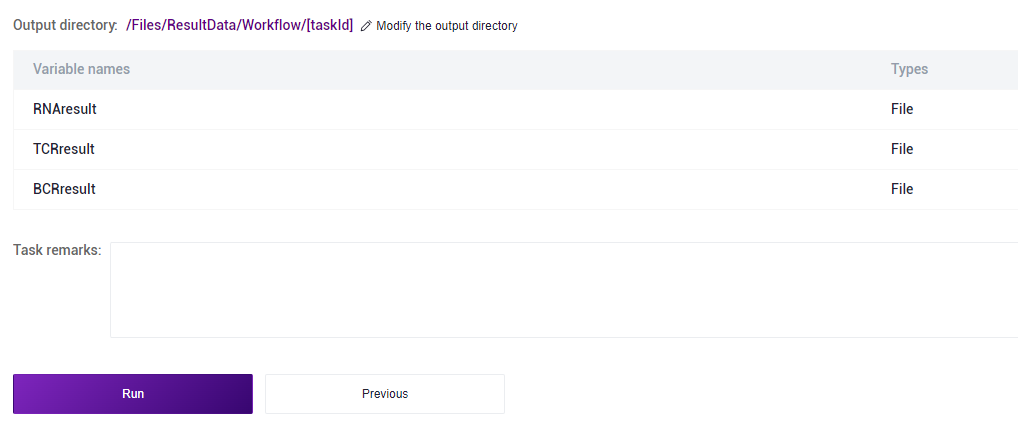
3.3 Use Scenario 2: Form Delivery
The operation consists of five steps: uploading data, constructing a reference genome (optional), downloading a sample template, filling in and importing the sample template, and initiating the analysis. After importing the sample template, you can run tasks in batches. When the customer sees that the task status is![]() When completed, it means that the task has been completed and you can view the report section (see section 3.4 for details).
When completed, it means that the task has been completed and you can view the report section (see section 3.4 for details).
3.3.1 Step 1: Upload Data
It is consistent with step 1 of Scenario 1 (manual delivery) (see Step 1 of 3.2.1: upload data for details).
3.3.2 Step 2: Build a reference genome (optional)
It is consistent with step 2 of Scenario 1 (manual delivery) (see Step 2 of 3.2.2: Construction of reference genome for details).
3.3.3 Step 3: Sample Information Entry Form Download
Click [Data] in the navigation bar, select [Table]-[Download] (as shown in Figure 3-14), click [Data model template], and select the scVDJ-seq template to Download:
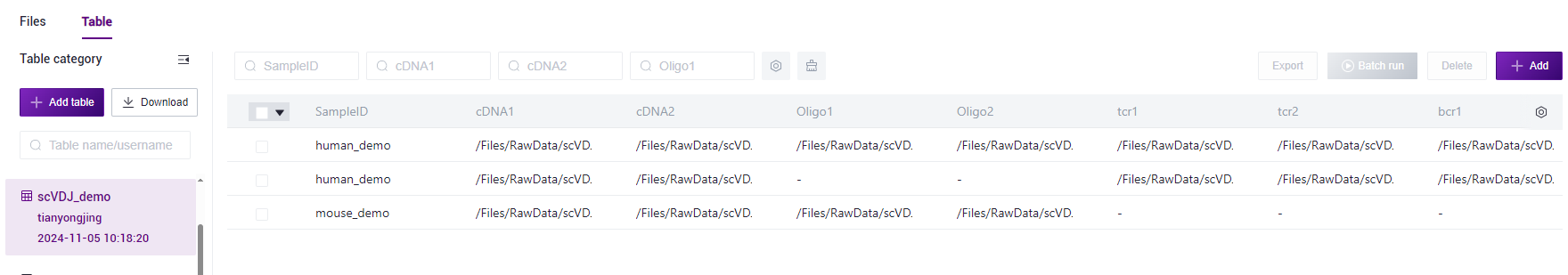
Figure 3-14 scVDJ-seq Sample Information Template Download Navigation After opening, the scVDJ-seq sample template Excel is shown in Figure 3-15:

3.3.4 Step 4: Import sample information
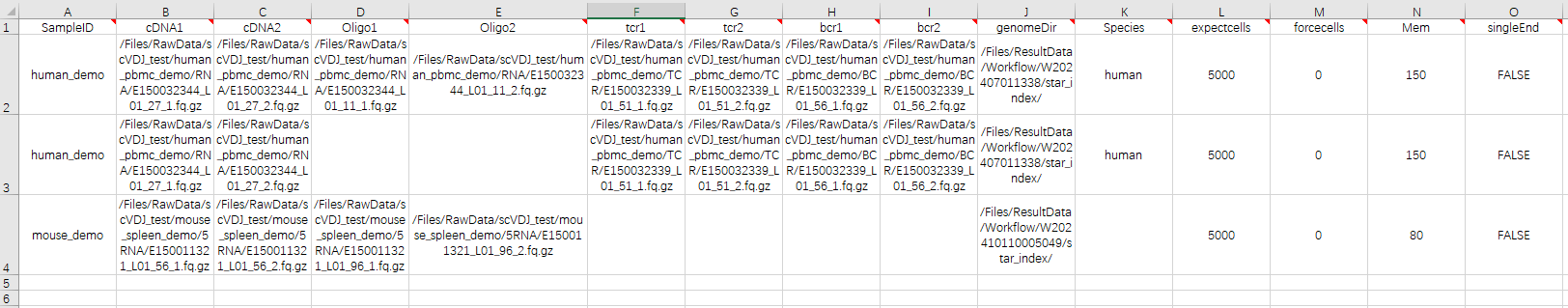
- Under the conditions of this usage scenario, the sample import form needs to fill in the worksheet (Figure 3-15). This scenario indicates that the sample data that has been sequenced is directly entered into the analysis after being imported into the form.
Note for Excel:
[1] The import file path must already exist on the cloud platform. If no file exists, no task is generated.
[2] In the template, input fields except tcr1, tcr2, bcr1, bcr2, Species, expectcells, and forcecells are required. In the import data, the required field must not be empty.
[3] SampleID in Excel must be unique.
[4] cells cannot be merged in Excel, and there cannot be spaces or special characters before and after cell contents.
[5] Analysis sample entry (Figure 3-16):
Tips
SampleID: the name of the sample, which is the same as the Entity ID by default;
cDNA: R1 and R2 end sequences in fastq format of cDNA library;
Oligo: the R1 and R2 end sequences of the oligo Library in fastq format;
tcr: Optional. The R1 and R2 end sequences of the tcr Library in fastq format, if not filled in, the TCR library will not be analyzed;
bcr: Optional. The R1 and R2 end sequences of bcr Library in fastq format, if not filled in, the BCR library will not be analyzed;
genomeDir: a reference genome file for 5-terminal transcriptome analysis, such as self-construction in step 2 (3.2.2 Construction of reference genome), select the star-index file in the Task ID folder;
Species: the reference database for VDJ analysis. It only supports human or mouse. Only human or mouse can be filled in. If other strings are filled in, TCR and BCR Library analysis will not be performed. If tcr or bcr files are not entered, this parameter will not take effect and may not be filled in;
expectcells: the desired number of cells, the number of cells can be selected according to experience;
forcecells: force the number of beads to be selected, and force the number of beads to be intercepted according to the sudden drop chart; If it is 0, it will not participate in the calculation and retain all cells;
mem: memory size required for running. The default value is 60g. If you need to run VDJ analysis, you need to run a large memory, we recommend that you set at least 100g.
singleEnd: indicates whether to use single-ended data Assembly. The default value is not used.
After configuring the analysis sample of the sample template and entering the worksheet, return to the [Data] interface and click [Table]-[+ Add table] (Figure 3-17):

Figure 3-17 Sample Information Import Step 1 Click [Click to upload/ Drop here] to browse and select the form with filled sample information, and Click [confirm] (Figure 3-18). After uploading, the file will be displayed in the target folder:
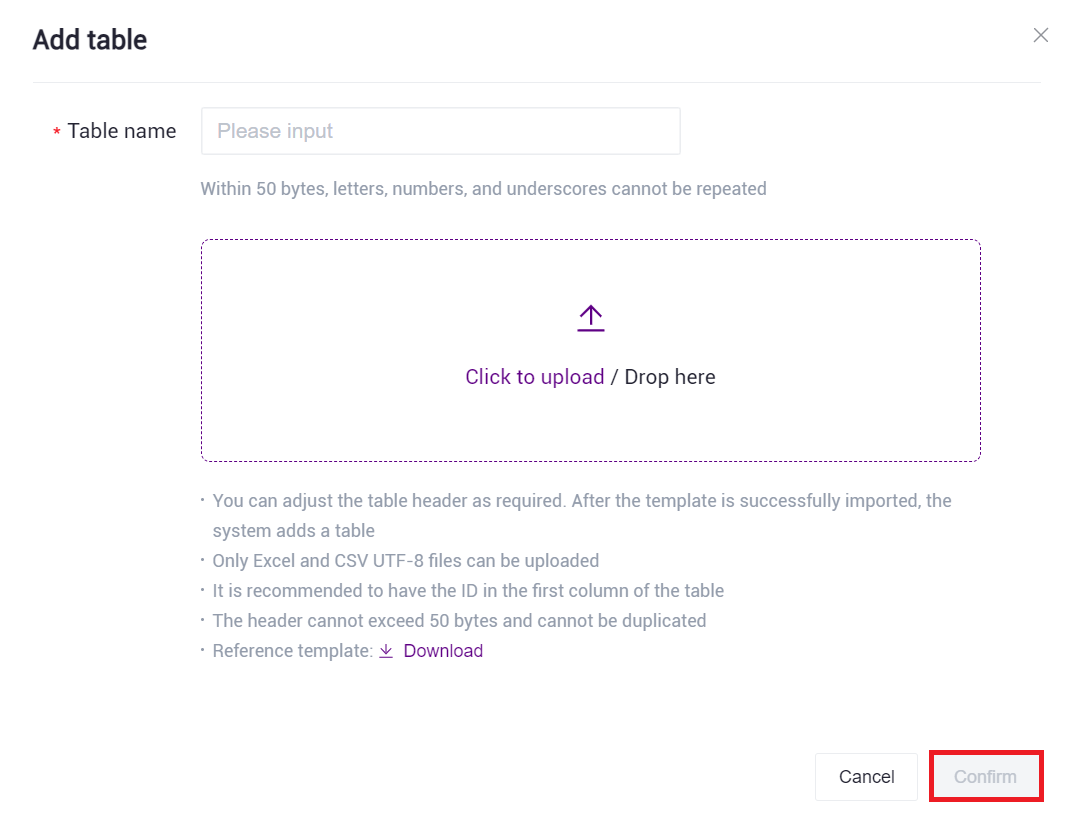
Figure 3-18 Sample Information Import Step 2 Click [Workflow] in the navigation bar to enter the process analysis page, enter scVDJ-seq in the search box, and click [Run] (Figure 3-19):
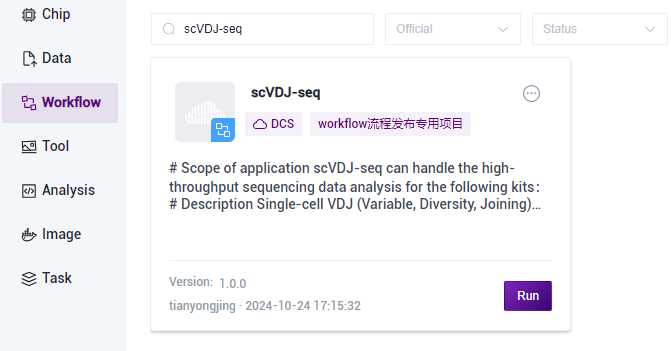
Figure 3-19 Sample Information Import Step 3 选择Run workflow(s),点击 Please select table
 Select the table imported in 3) of this section, select the required row, and click [Next] (Figure 3-20):
Select the table imported in 3) of this section, select the required row, and click [Next] (Figure 3-20):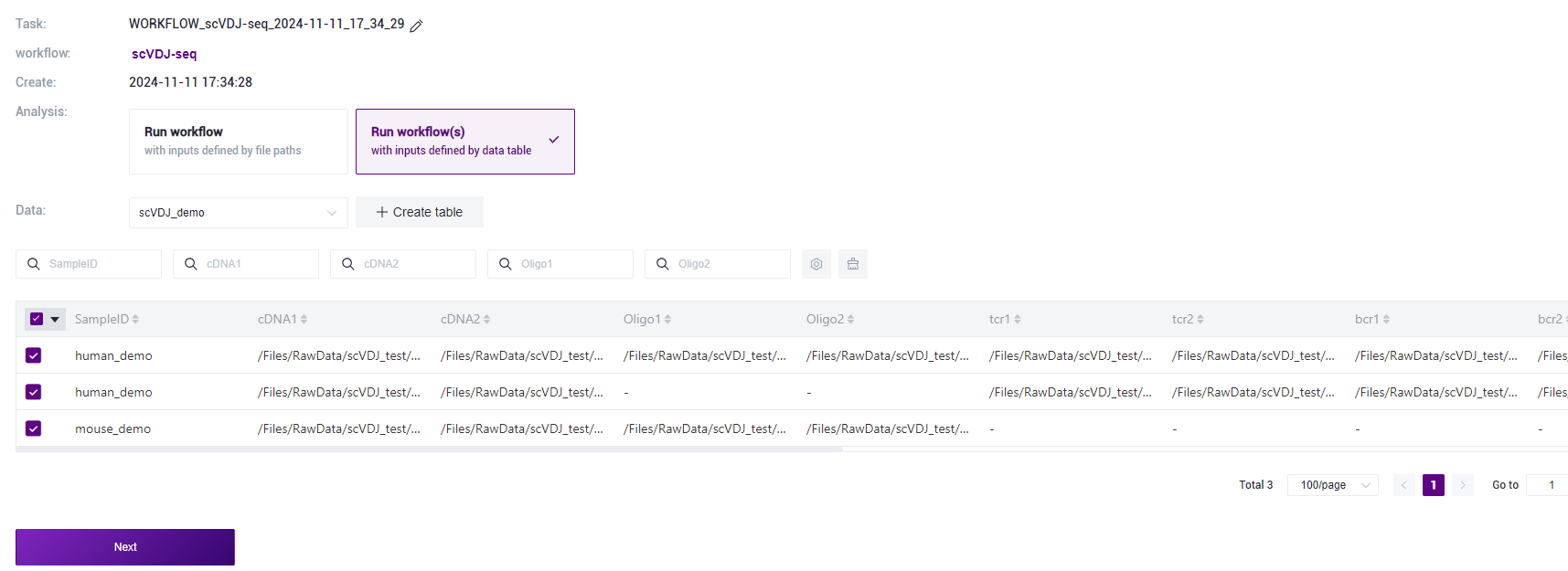
Figure 3-20 Sample Information Import Step 4 Click at Values
 And select the corresponding value, such as cDNA select {cDNA2}, pay attention to the order of selection (as shown in Figure 3-21):
And select the corresponding value, such as cDNA select {cDNA2}, pay attention to the order of selection (as shown in Figure 3-21):
Figure 3-21 Sample Information Import Step 5 Enter the sample information and click [Next] after completion to ensure that the parameter settings are correct (as shown in Figure 3-22):
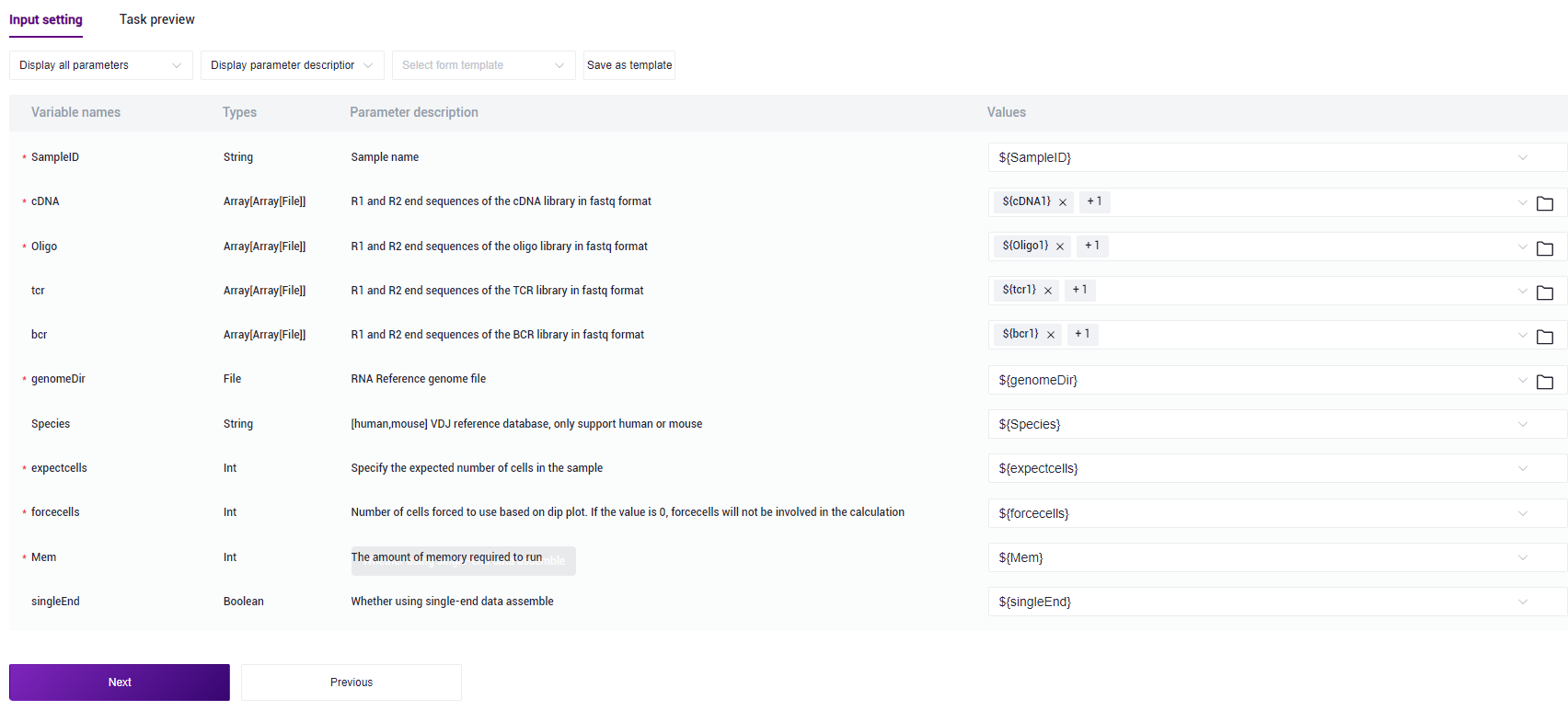
3.3.5 Step 5: Start Analysis
- Click [Run] to start the analysis (Figure 3-23):
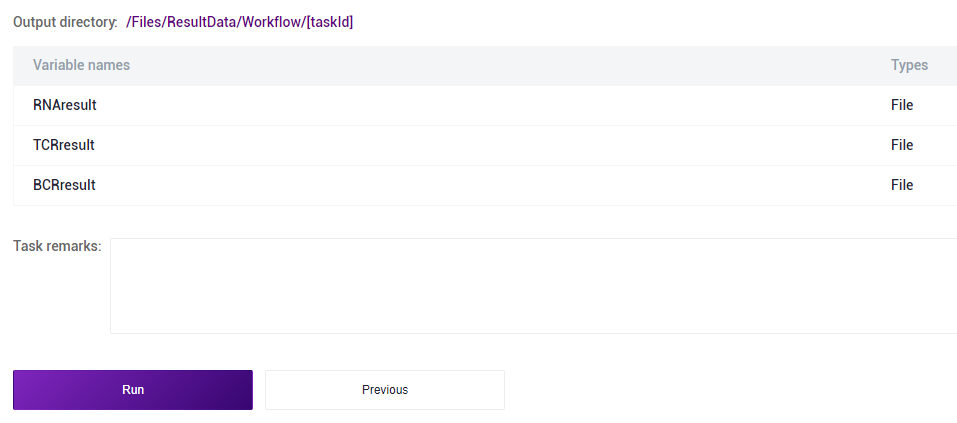
Report viewing and result file download
Report View
- Click the navigation bar [Task], when the Task status is displayed
 When completed, it means that the task has been completed. Click [Detail] to view the result part (as shown in Figure 3-24):
When completed, it means that the task has been completed. Click [Detail] to view the result part (as shown in Figure 3-24):
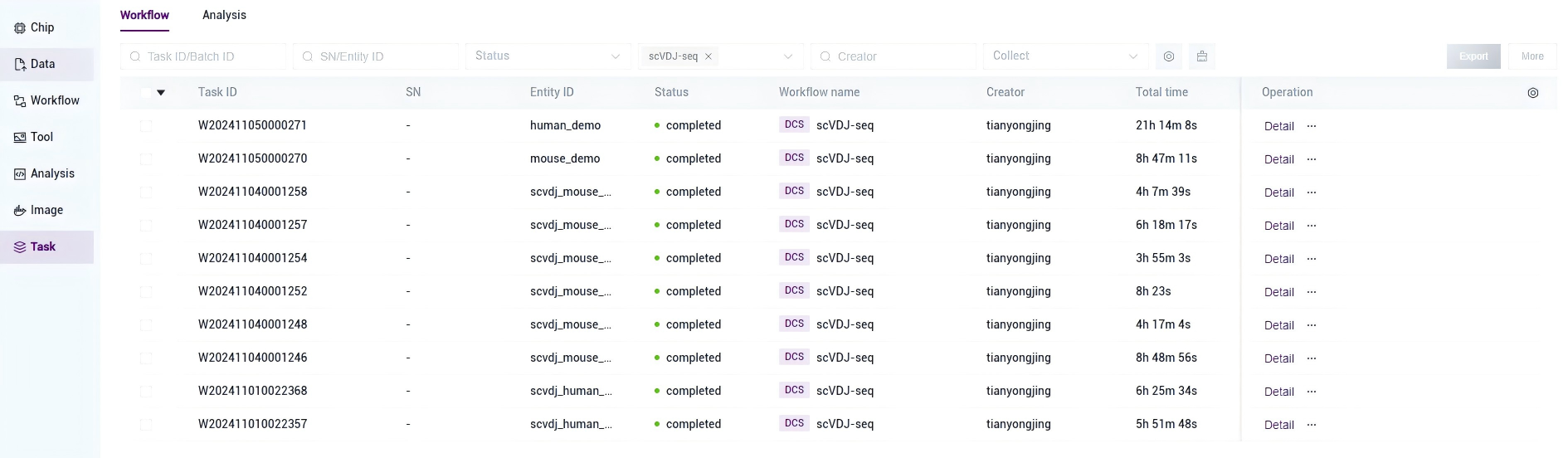
Report Download
Click the navigation bar [Data] to enter the Data management page, search according to the Task ID of the Task, and click to enter the Task ID folder (as shown in Fig. 3-25):
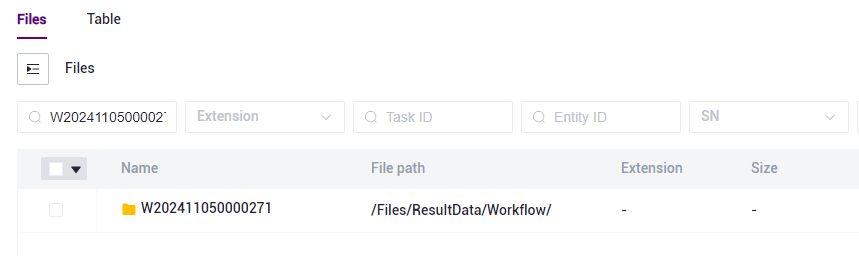
Figure 3-25 Report Download Step 1 If you need RNA or TCR or BCR to analyze the corresponding report, you need to click to enter the RNA or TCR or BCR folder respectively (as shown in Figure 3-26):
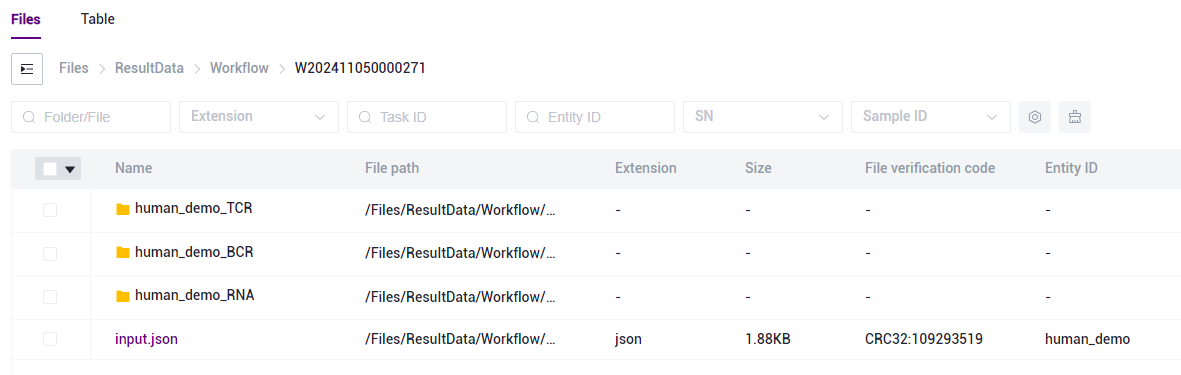
Figure 3-26 Report Download Step 2 Click to enter the output folder (as shown in Figure 3-26):
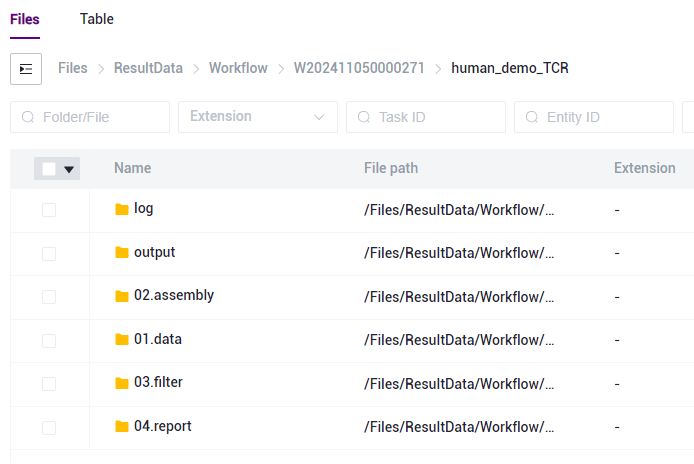
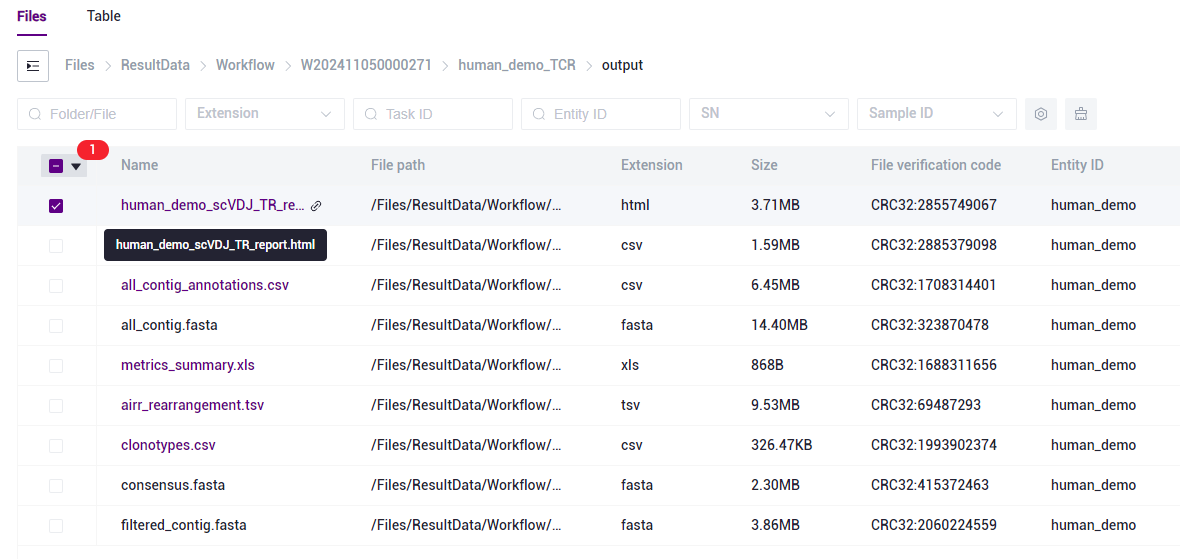
Figure 3-26 Report Download Step 2 Select the report.html file and click [Download]-[Raysync download] (as shown in Figure 3-27):
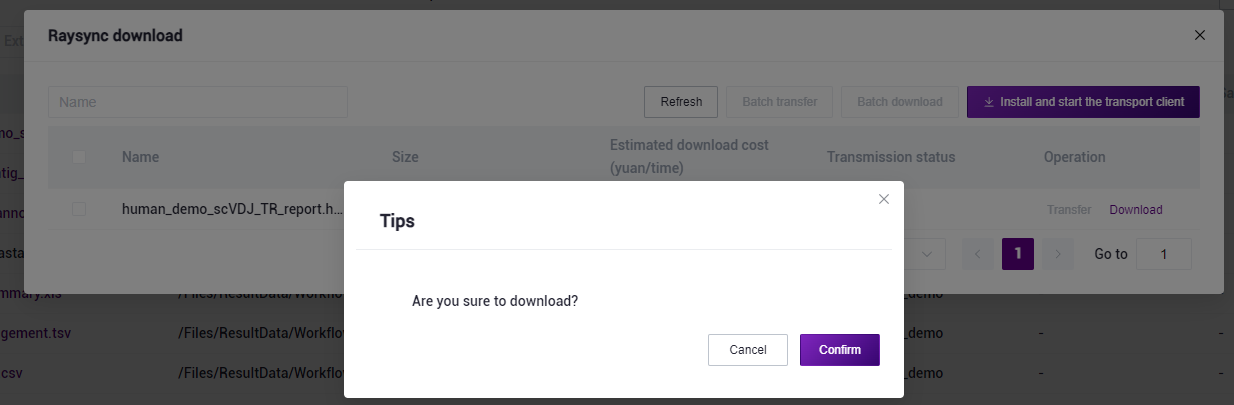
- Click [Download]-[Confirm], select the target directory and Download the report (as shown in Figure 3-28):
Chapter 4 Results Presentation
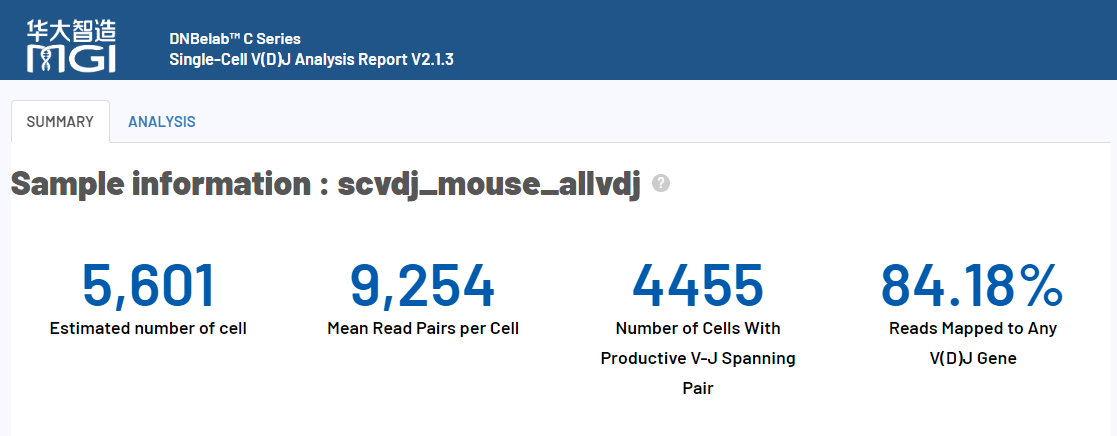
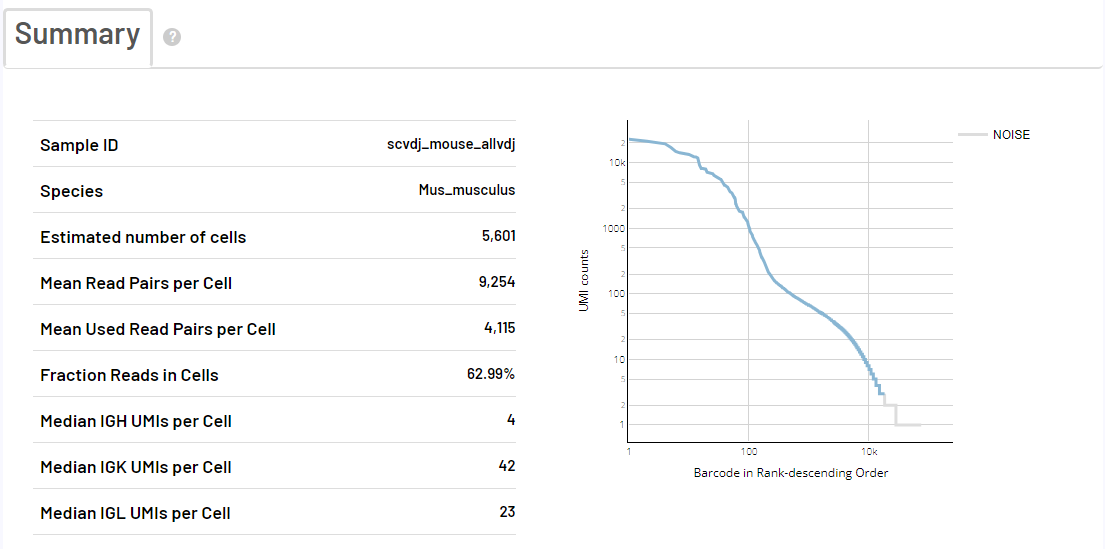
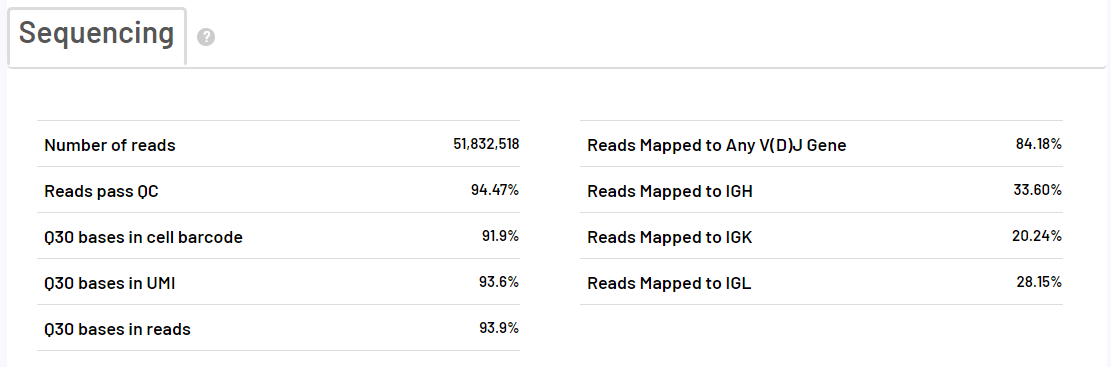
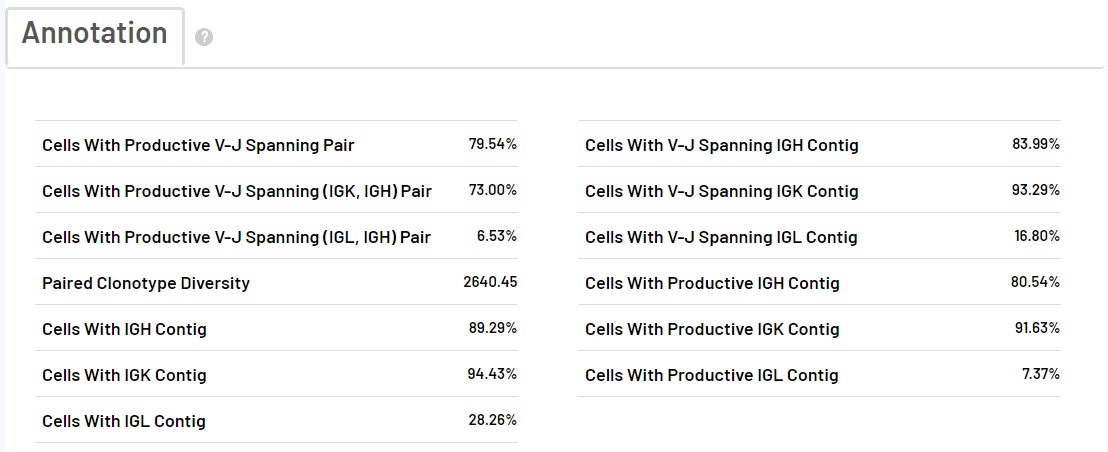
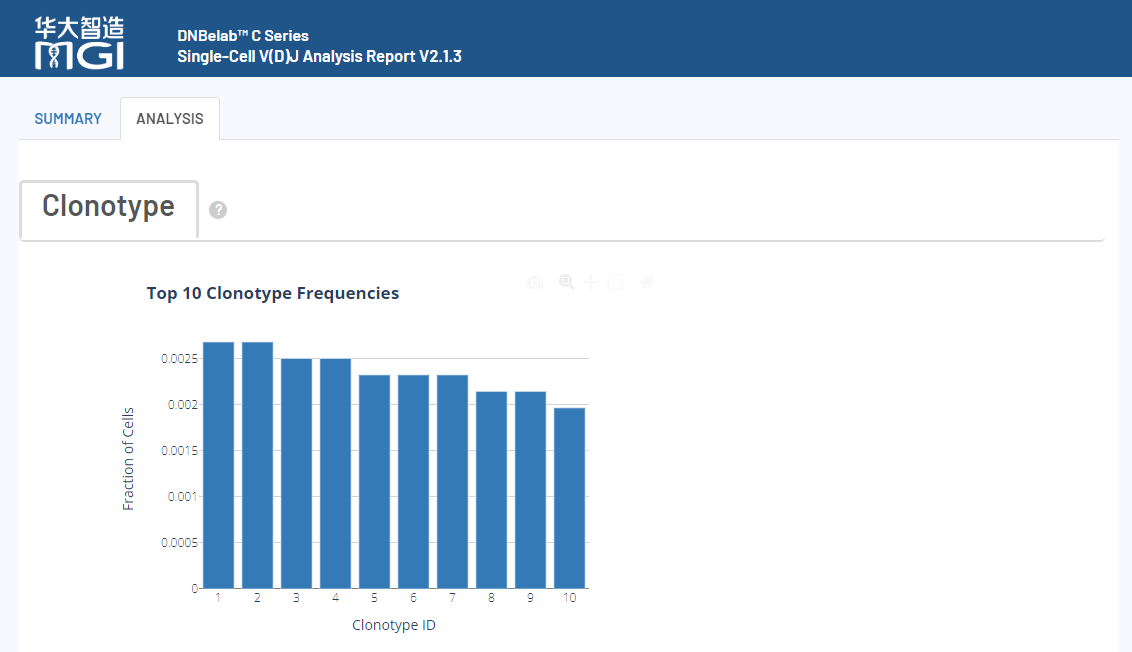
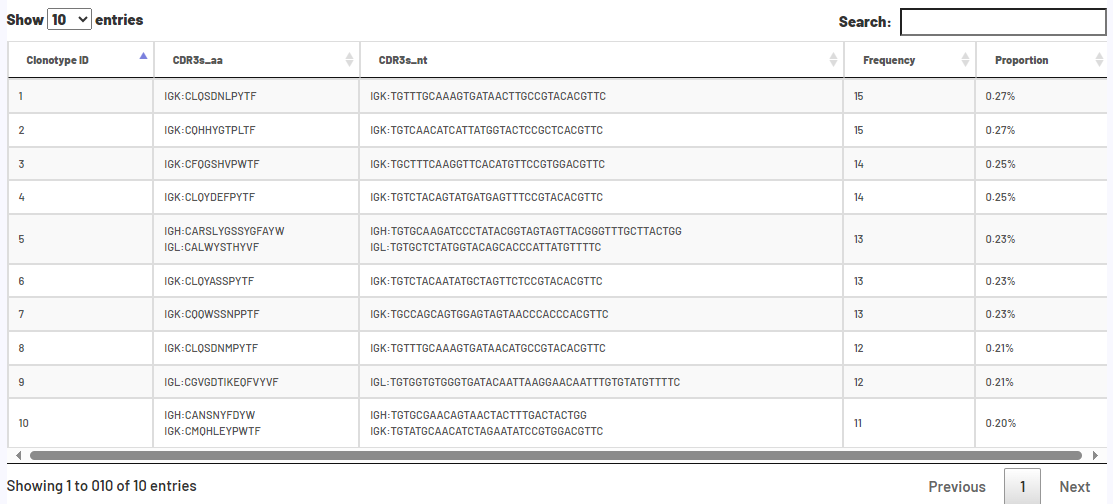
scVDJ-seq Sample Report Presentation
The report of the 5-terminal single-cell transcriptome is the same as the report of the scRNA-seq_v3 process. The following takes BCR analysis as an example to show only the report of VDJ analysis.